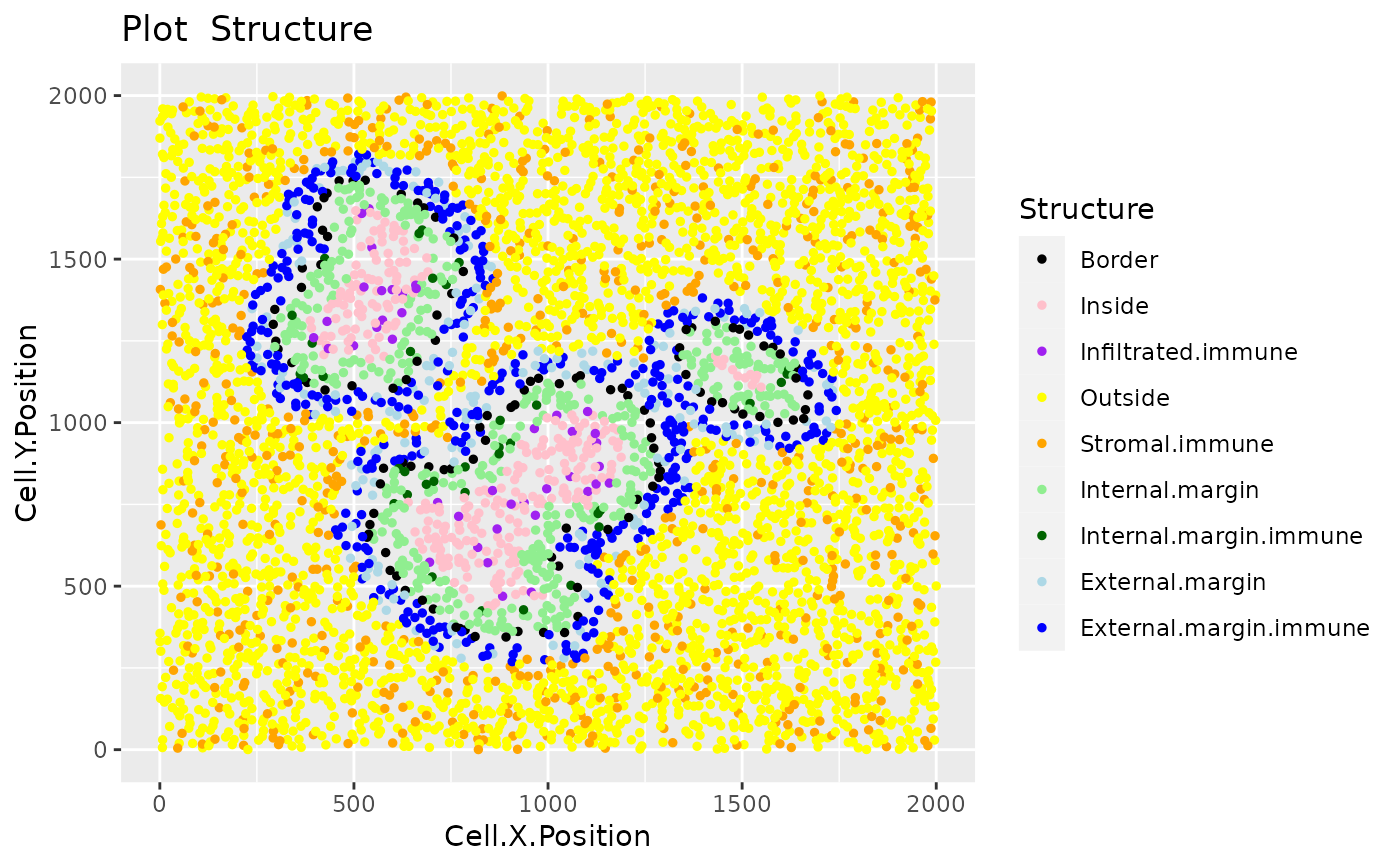
After identifying the bordering cells of tumour regions and calculating the distances of each cell to the tumour bordering cells, this function further identifies the cells that are located in the inside and outside of the tumour regions, and in the internal and external tumour margins. It also identifies the immune cells that are infiltrated, stromal, internal margin or external margin immune cells.
Usage
define_structure(
sce_object,
names_of_immune_cells,
feature_colname = "Cell.Type",
n_margin_layers = 5
)Arguments
- sce_object
SingleCellExperiment object whose metadata has the information of tumour bordering cells and cell distances to tumour border (has columns "Region" and "Distance.To.Border").
- names_of_immune_cells
String Vector of the names of immune cells.
- feature_colname
String Specifying the column that contains the names of the immune cells.
- n_margin_layers
Integer. The number of layers of cells that compose the internal/external tumour margins.
Examples
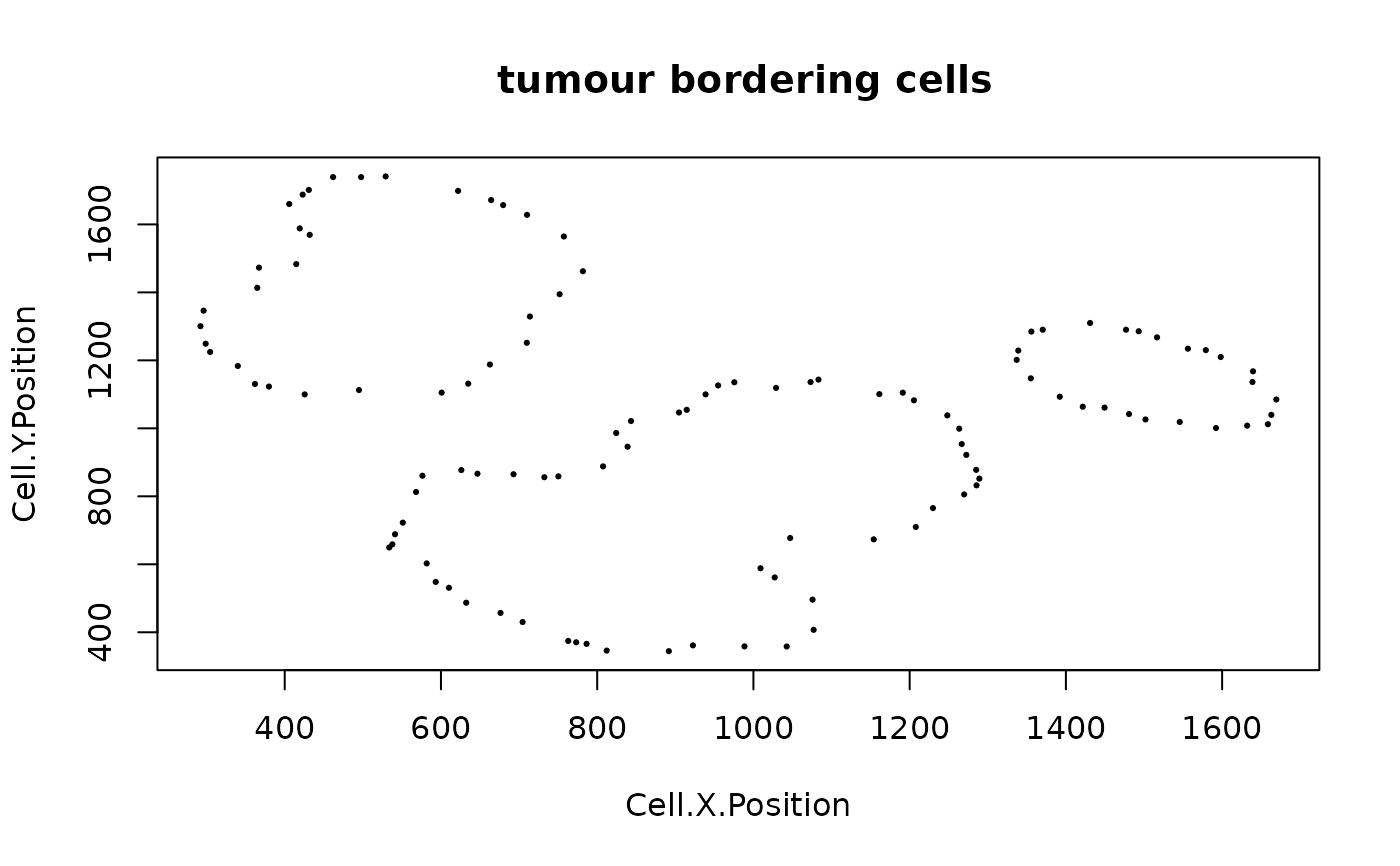
sce_border <- identify_bordering_cells(SPIAT::defined_image,
reference_cell = "Tumour", feature_colname = "Cell.Type", n_to_exclude = 10)
#> [1] "The alpha of Polygon is: 63.24375"
 sce_dist <- calculate_distance_to_tumour_margin(sce_border)
#> [1] "Markers had been selected in pair-wise distance calculation: "
#> [1] "Non-border" "Border"
sce_structure <- define_structure(sce_dist,
names_of_immune_cells = c("Immune1","Immune2","Immune3"),
feature_colname = "Cell.Type", n_margin_layers = 5)
plot_cell_categories(sce_structure, feature_colname = "Structure")
sce_dist <- calculate_distance_to_tumour_margin(sce_border)
#> [1] "Markers had been selected in pair-wise distance calculation: "
#> [1] "Non-border" "Border"
sce_structure <- define_structure(sce_dist,
names_of_immune_cells = c("Immune1","Immune2","Immune3"),
feature_colname = "Cell.Type", n_margin_layers = 5)
plot_cell_categories(sce_structure, feature_colname = "Structure")