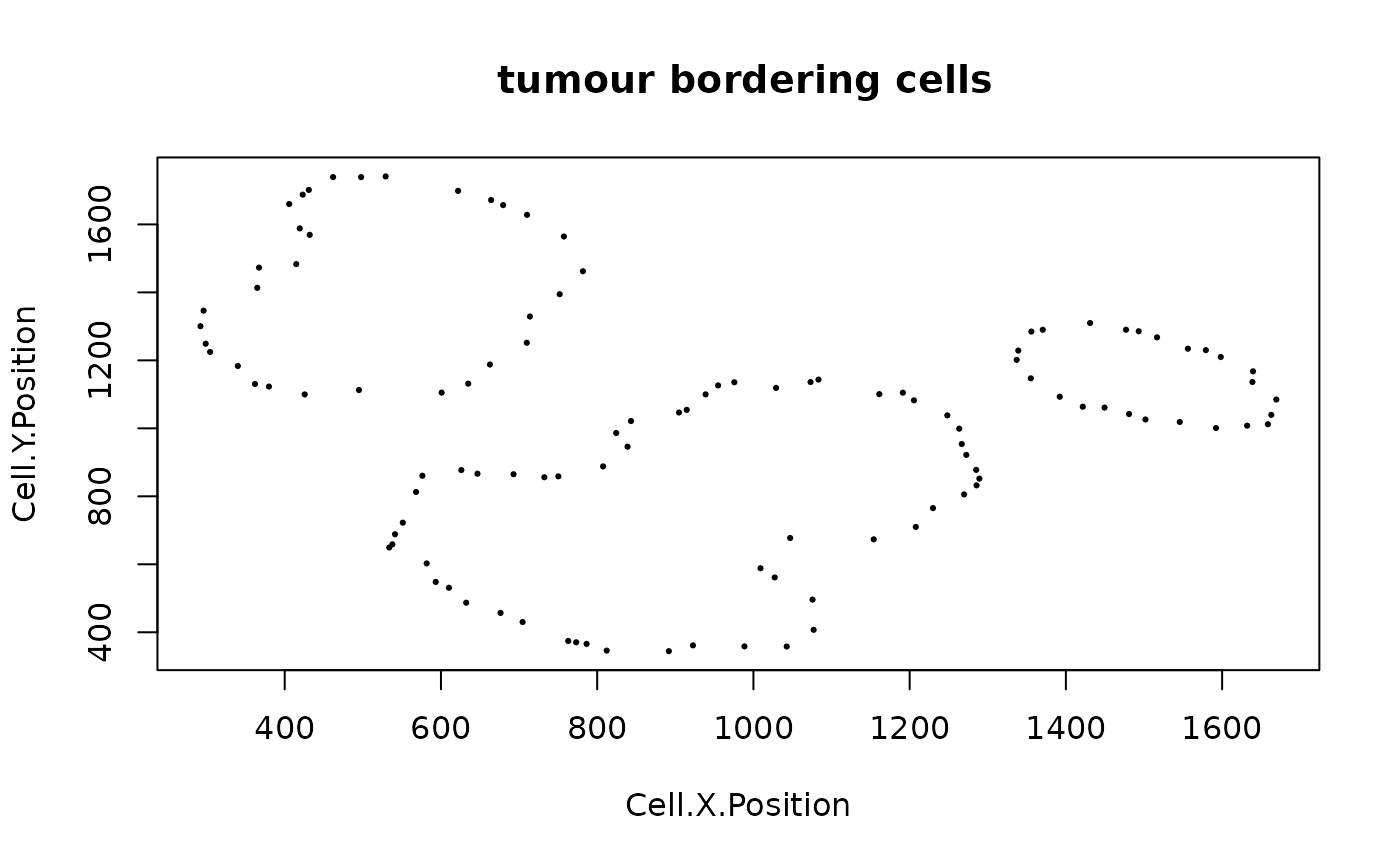
Identify the cells bordering a group of cells of a particular phenotype.
Usage
identify_bordering_cells(
sce_object,
reference_cell,
feature_colname = "Cell.Type",
ahull_alpha = NULL,
n_of_polygons = 1,
draw = FALSE,
n_to_exclude = 10
)Arguments
- sce_object
SingleCellExperiment object in the form of the output of
format_image_to_sce.- reference_cell
String. Cells of this cell type will be used for border detection.
- feature_colname
String that speficies the column of `reference_cell`.
- ahull_alpha
Number specifying the parameter for the alpha hull algorithm. The larger the number, the more cells will be included in one cell cluster.
- n_of_polygons
Integer specifying the number of tumour regions defined by user.
- draw
Boolean if user chooses to manually draw the tumour area or not. Default is False.
- n_to_exclude
Integer. Clusters with cell count under this number will be deleted.
Details
The bordering cell detection algorithm is based on computing an alpha hull (Hemmer et al., 2020), a generalization of convex hull (Green and Silverman, 1979). The cells detected to be on the alpha hull are identified as the bordering cells.
Examples
sce_border <- identify_bordering_cells(SPIAT::defined_image, reference_cell = "Tumour",
feature_colname = "Cell.Type", n_to_exclude = 10)
#> [1] "The alpha of Polygon is: 63.24375"